Registration of diffusion tensor images
We consider the problem of registering
diffusion tensor images under various local deformation models.
Existing methods for DTI registration are computationally intensive and good initialization is critical.
Our objective is to develop simple linear
registration algorithms that can be used for initializing computationally
intensive methods.
In [1,2], we proposed an algebraic method for
the registration of DTI data.
We derive the so-called Diffusion Tensor Constancy Constraint, a
generalization of the Brightness Constancy Constraint for 2D images to
diffusion tensor data. Under the standard Euclidean metric in SPSD(3), we show that
for various local deformation models, such as translational, rigid, and affine,
together with the finite strain reorientation scheme as shown in Figure 1, the DTCC leads
to a linear relationship between the parameters of the
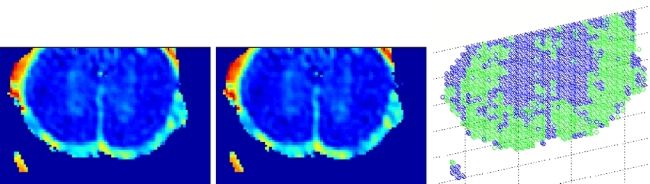
deformation, the DT data and its first order partial derivatives. Figure 2 shows the results of our registration algorithm using a multi-scale implementation.
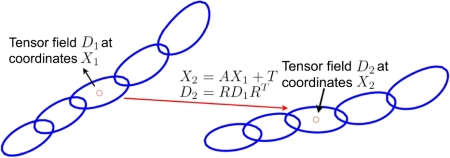
Figure 1: Registration of two DTI data sets under affine deformation model with finite strain reorientation scheme.
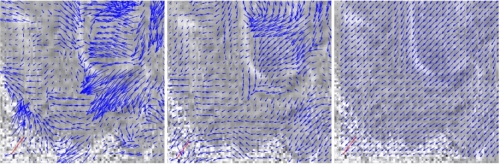
Figure 2: (Left and center) Deformation at scale 4 and 2. (Right) Final deformation.

Figure 1: Registration of two DTI data sets under affine deformation model with finite strain reorientation scheme.

Figure 2: (Left and center) Deformation at scale 4 and 2. (Right) Final deformation.